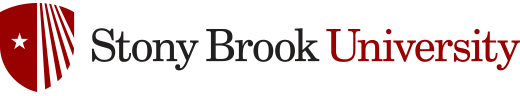Sterylglucosidases as Antifungal Targets
Invasive fungal infections by fungi such as C. neoformans and A. fumigatus have devastating effects on human health and quality of life. Exposure to both C. neoformans and A. fumigatus is quite common through air-borne spores, and immunocompetent individuals mount an effective immune response that results in small granulomas within the lung that house dormant spores.1,2 People with compromised immune systems are not able to produce an suitable immune response, which allows these fungi to grow and ultimately progress to their symptomatic diseases; cryptococcosis and aspergillosis, respectively. Aspergillosis and cryptococcosis have unique disease pathways, with C. neoformans propagating from the lungs to the central nervous system, causing inflammation and lethal meningoencephalitis, while A. fumigatus remains within the lungs and grows into large fungal tumors. Despite these differences, both C. neoformans and A. fumigatus require sterylglucosidase activity to avoid the mammalian immune response. Sterylglucosidase - a fungi-specific enzyme - hydrolyzes sterylglucosides, such as Ergosterol-3β-D-glucoside (ErgGlc) shown in Figure 1.
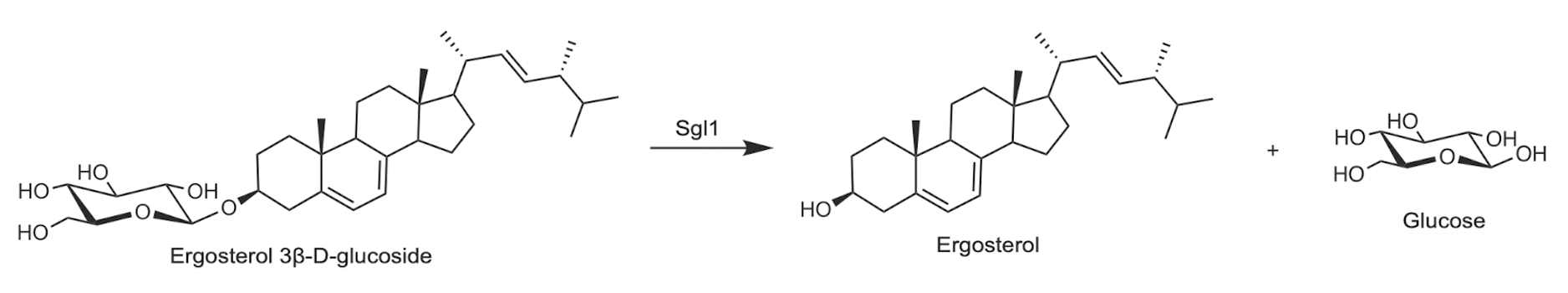
Figure 1. ErgGlc hydrolysis by sterylglucosidase
Sterylglucosidase expressed by both C. neoformans (Sgl1) and A. fumigatus (SglA) have been validated as anti-fungal drug targets. As shown in Figure 17, both wild type (WT) and reconstituted fungal cells (∆sgl+SGL) show steep survival curves, while the genetic ablation (∆sgl) hovers at 100% survival.3,4 This definitively shows that sterylglucosidase activity is required for pathogenicity of both A. fumigatus and C. neoformans.
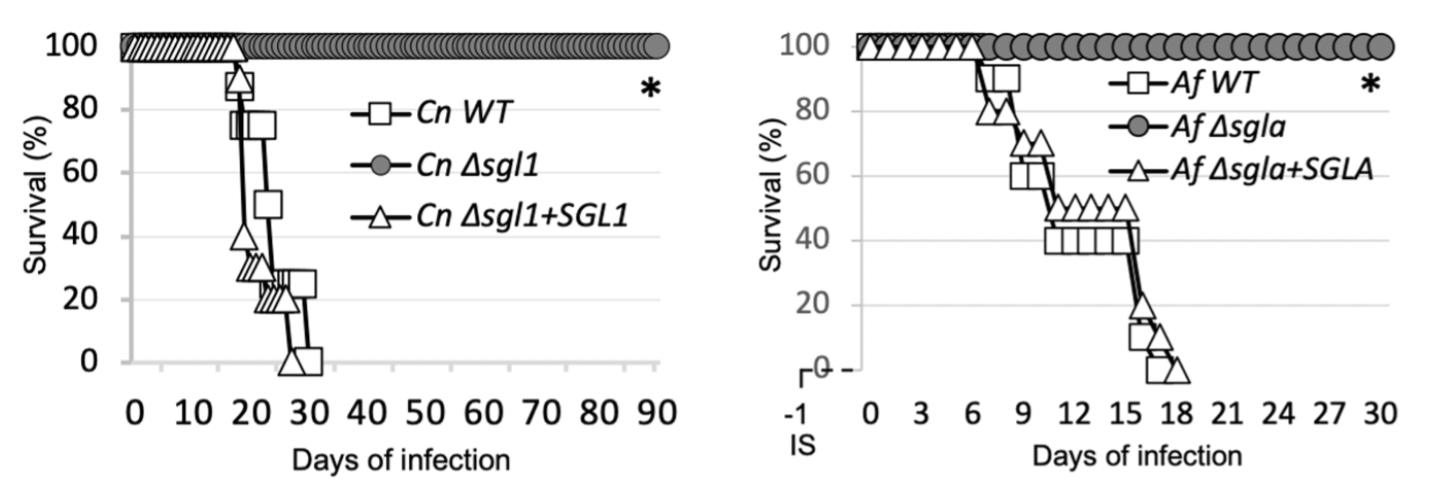
Figure 2. Sgl1 (left) and SglA (right) target validation.
Erg-glc has not yet been successfully co-crystallized with either wild type or catalytically inactive Sgl1-E520S. However, computational approaches have been successful in predicting the binding mode of lipid substrates to lipid-modifying enzymes with well-defined hydrophobic pockets like Sgl1. To understand the mechanistic basis for erg-glc substrate recognition and hydrolysis, we used a flexible ligand docking protocol to dock erg-gcl in the Y-shaped pocket of Sgl1. Erg-glc was docked with the glucose moiety in position for nucleophilic attack by the catalytic glutamate residue Glu520 and the ergosterol moiety was surrounded by hydrophobic residues in the buried hydrophobic arm of the Y-shaped pocket. Glucose recognition was mediated by several hydrogen-bond contacts with the residues Lys47, His142, Asn269, Glu270, Trp570, and Glu587. Based on the predicted binding pose of erg-glc, we propose that Sgl1 uses a canonical reaction mechanism for erg-glc hydrolysis and a series of polar and non-polar interactions to generate affinity for the glucose and ergosterol moieties of erg-glc. To test this hypothesis, we generated 12 single point mutants of key residues putatively involved in erg-glc recognition and catalysis and tested their activity towards erg-glc and the artificial substrate resorufin-3β-d-glucopyranoside (res-glp). Consistent with our predictions and a prior study of a bacterial endoglucoceramidase, Alanine point mutations of residues predicted to directly bind to the glucose moiety (Lys47, His142, Asp144, Tyr453, Trp570, and Glu587), as well as point mutations of the catalytic glutamic acids Glu270 and Glu520, either greatly reduced activity or completely inactivated Sgl1. Except for Glu270, which is the catalytic glutamate that initiates catalysis, all point mutants displayed some residual activity towards either erg-glc or res-glp. This suggests the observed reduction of activity is not due to protein misfolding, but rather to the importance of these residues in substrate recognition and/or catalysis.

Figure 3. ErgGlc docked pose (left) and mutation study (right)
A high-throughput screening of 50,000 compounds against Sgl1 and SglA were conducted and identified a number of hit compounds. First, the Sgl1 screen identified Hit 1, Hit 9 and Hit 15, shown in Figure 4. Each hit both inhibited Sgl1 and successfully caused the accumulation of ErgGlc, which is the phenotypic goal of Sgl inhibition.

Figure 4. HTS against Sgl1.
Crystal structures of Hit 1 and Hit 9 bound to Sgl1 were resolved through X-ray crystallography, and further supported the identification of the catalytic site, as shown in Figure 5 and 6.
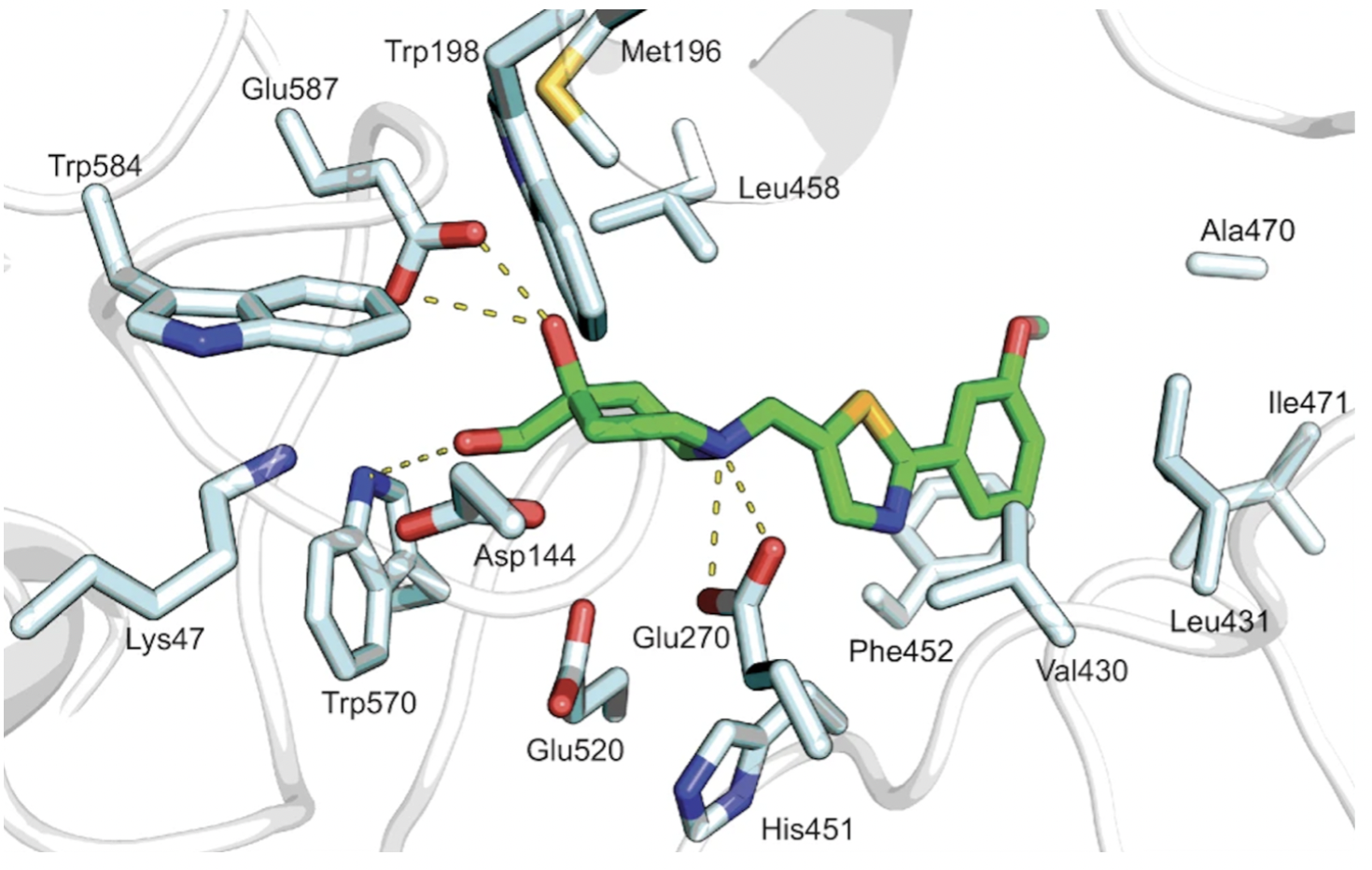
Figure 5. Hit 1-Sgl1 crystal structure (PDB: 7LPP)
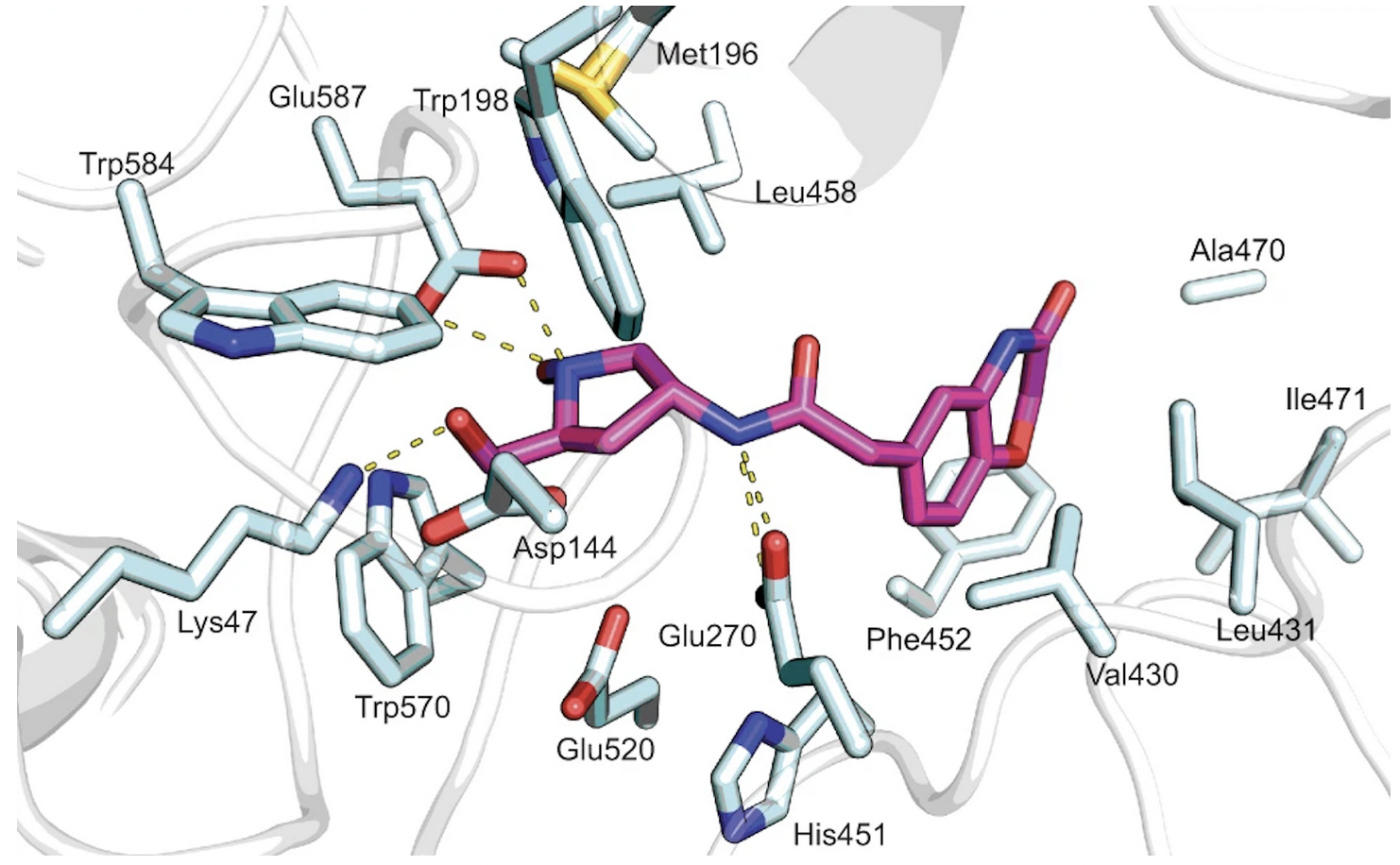
Figure 6. Hit 9-Sgl1 crystal structure (PDB: 7LPQ)
The SglA high-throughput screening identified two unique hit compounds, as shown in Figure 7, although no cocrystal structure has been resolved to date. The two hits identified in the HTS campaign (hit B and hit C) were submitted to a two-step validation. First, the ability to inhibit the hydrolysis of ErgGlc in vitro was tested. Dose-dependent inhibition of ErgGlc hydrolysis by SglA was observed for both compounds, with IC50s in the low micromolar range. In the second step, the capacity of these SglA inhibitors to promote the accumulation of ErgGlc inside live Af wild-type cells was evaluated. Upon treatment of Af wild-type cells for 48 h, we observed a significant and dose-dependent increase in the intracellular concentration of ErgGlc, with the maximal accumulation of ErgGlc at the highest concentration for both compounds. Both hits promoted a higher accumulation of ErgGlc at the highest concentrations tested (100 and 500 μM). Hits B and C differed significantly from the control at 100 and 500 μM concentrations after 48 h of treatment. Notably, both hits B and C at 500 μM led to an accumulation of ErgGlc similar to that seen with the Af Δsgla strain, where SglA is genetically deleted.

Figure 7. HTS against SglA
Although both hit compounds were promising, further exploration of compounds similar to Hit B revealed a more potent compound, Hit B7 (Figure 8). Hit B7 more effectively inhibited SglA than any other Hit B analog or Hit B itself, leading it to be the lead inhibitor against SglA.

Figure 8. Hit compounds from high throughput screening of SglA.
Finally, Hit B7 was evaluated both in vitro and in vivo (Figure 9). Since B7 showed a better affinity for SglA than hit B, we assessed the ability of this compound to promote the accumulation of ErgGlc and to delay hyphal elongation in the Af wild-type strain. B7 treatment for 48 h of wild-type Af led to a significantly higher accumulation of ErgGlc than treatment with hit B at 100 μM. Moreover, this accumulation at 100 μM is similar to the accumulation observed in the mutant Δsgla strain. In addition, B7 treatment promotes a significant delay in hyphal growth, performing even better than hit B at 100 μM, resulting in hyphal lengths that are similar to those in the Δsgla strain. The toxicity of B7 in the mammalian cell line A549 was slightly worse than that of the parent compound, with an LD50 of 200 μM, compared to 350 μM for hit B.

Figure 9. Hit B7 evaluation
Using these data, we aim to use computer-aided drug design (CADD) to develop Sgl1 and SglA inhibitors to treat cryptococcosis and aspergillosis, respectively.
Related Publications
1. Kechichian, T. B.; Shea, J.; Del Poeta, M. Depletion of Alveolar Macrophages Decreases the Dissemination of a Glucosylceramide-Deficient Mutant of Cryptococcus Neoformans in Immunodeficient Mice. Infection and Immunity2007, 75 (10), 4792–4798. https://doi.org/10.1128/IAI.00587-07.
2. McQuiston, T.; Luberto, C.; Del Poeta, M. Role of Host Sphingosine Kinase 1 in the Lung Response against Cryptococcosis. Infection and Immunity2010, 78 (5), 2342–2352. https://doi.org/10.1128/IAI.01140-09.
3. Pereira de Sa, N.; Jayanetti, K.; Rendina, D.; Clement, T.; Soares Brauer, V.; Mota Fernandes, C.; Ojima, I.; Airola, M. V.; Del Poeta, M. Targeting Sterylglucosidase A to Treat Aspergillus Fumigatus Infections. mBio2023, 14 (2), e0033923. https://doi.org/10.1128/mbio.00339-23.
4. Rella, A.; Mor, V.; Farnoud, A. M.; Singh, A.; Shamseddine, A. A.; Ivanova, E.; Carpino, N.; Montagna, M. T.; Luberto, C.; Del Poeta, M. Role of Sterylglucosidase 1 (Sgl1) on the Pathogenicity of Cryptococcus Neoformans: Potential Applications for Vaccine Development. Frontiers in Microbiology2015, 6.
5. Pereira de Sa, N.; Taouil, A.; Kim, J.; Clement, T.; Hoffmann, R. M.; Burke, J. E.; Rizzo, R. C.; Ojima, I.; Del Poeta, M.; Airola, M. V. Structure and Inhibition of Cryptococcus Neoformans Sterylglucosidase to Develop Antifungal Agents. Nat Commun2021, 12 (1), 5885. https://doi.org/10.1038/s41467-021-26163-5.